
We recommend always using the latest version of Java. It should be possible to run PeptideShaker on almost any computer where Java 1.8 or newer is installed. To start using PeptideShaker, unzip the downloaded file, and double-click the PeptideShaker-X.Y.Z.jar file. For specific bug reports or issues please use the issues tracker.
#Nokoi peptideshaker free#
If you have any questions, suggestions or remarks, feel free to contact us via the Reshake PRIDE: re-analyze public datasets in PRIDE as if they were your own.Īll data can also easily be exported for follow up analysis in other tools.įor further help see the Bioinformatics for Proteomics Tutorial.QC Plots: examine the quality of the results with Quality Control plots.Validation: inspect and fine tune the validation process.GO Enrichment: perform GO enrichment and find enriched GO terms in your dataset.3D Structure: map the detected peptides and modifications onto corresponding PDB structures.Modifications: get a detailed view of the post-translational modifications in the dataset.Fractions: inspect from which fractions proteins and peptides are likely to come from.Spectrum IDs: compare the search engine performance and see how the search engine results are combined.Overview: get a simple yet detailed overview of all the proteins, peptides and PSMs in your dataset.PeptideShaker currently supports nine different analysis tasks: It can be used on local data as well as on data sets deposited to the ProteomeXchange PRIDE repository. PeptideShaker can be used in command line and comes with rich visualization capabilities to navigate the results. PeptideShaker aggregates the results in a single identification set, annotates spectra, computes a consensus score, maps sequences and performs protein inference, scores post-translational modification localization, runs statistical validation, quality control, and annotates the results using multiple sources of information like Gene Ontology, UniProt and Ensembl annotation, and protein structures.
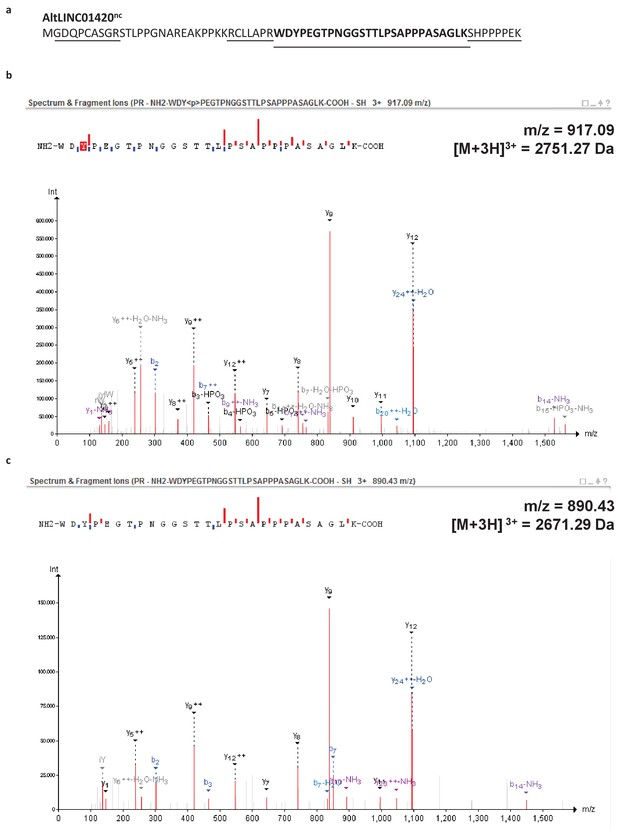
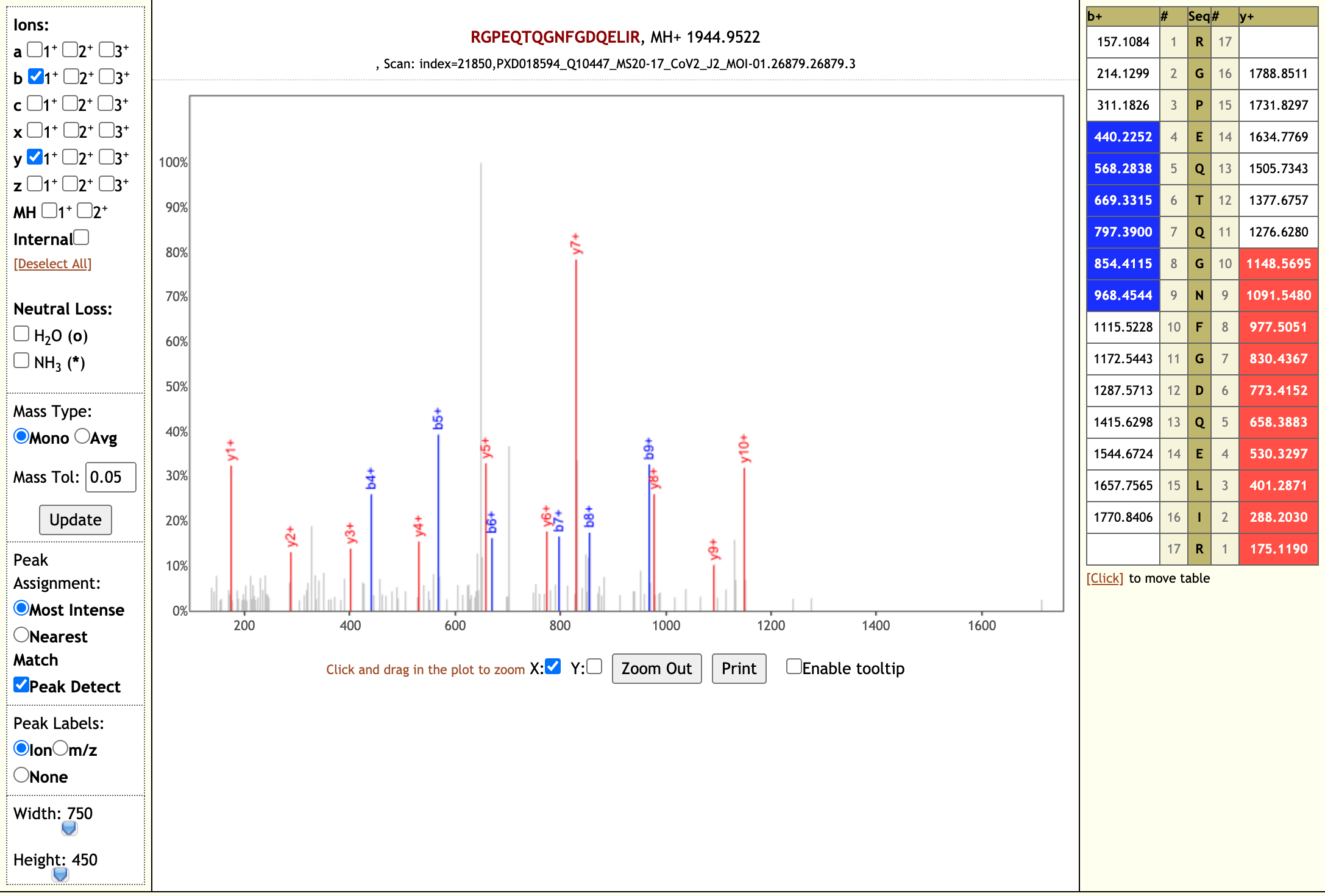
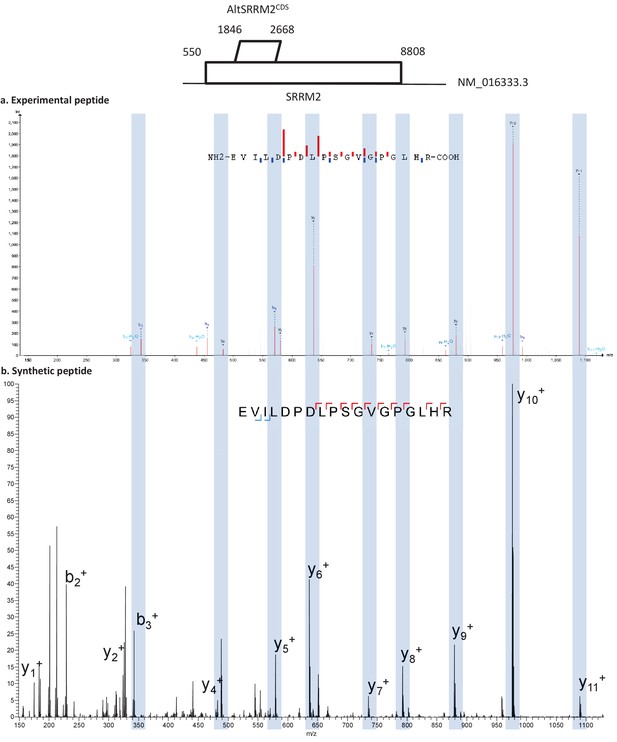
PeptideShaker is a search engine independent platform for interpretation of proteomics identification results from multiple search and de novo engines, currently supporting X!Tandem, MS-GF+, MS Amanda, OMSSA, MyriMatch, Comet, Tide, Mascot, Andromeda, MetaMorpheus, Novor, DirecTag and mzIdentML.
#Nokoi peptideshaker full size#
(Click on an image to see the full size version) If you use PeptideShaker as part of a publication please include this reference.


 0 kommentar(er)
0 kommentar(er)
